How can wastewater predict the future?
In healthcare, antimicrobial resistance (AMR) is a constantly growing issue – but it’s not a new one. For several years, scientists have cautioned against the overuse of prescription antibiotics and petitioned for increased surveillance of AMR within hospitals, clinics and throughout the general population. Yet, AMR continues to increase in scope and severity, despite efforts to implement antibiotic stewardship practices. This is partially due to the COVID-19 pandemic, during which most government efforts were focused on addressing the threat of SARS-CoV-2. With some of the pandemic-related urgency subsiding, the threat of AMR once again looms over public health officials. AMR didn’t stop because we were no longer monitoring it – it got worse.
Current efforts to address AMR
Often, plans for combating AMR hinge on antibiotic stewardship. While these practices help to curb the issue, they rely on accurate identification of the etiological organism – a process which may take a long time. This can be an issue, as most patients wait to go to the hospital until they are experiencing severe, sometimes life-threatening disease. Modern molecular diagnostic instruments have partially addressed this issue by analyzing patient samples in less time, but they are still subject to error. Without proper verification of the analysis, these instruments can quickly provide incorrect patient results, prompting inappropriate antibiotic use and possibly further perpetuating AMR.
MDx-Chex® for BCID2, MDx-Chex® for BC-GP and MDx-Chex® for BC-GN are patient-like full process controls designed to validate the entire analytical process of the BCID2, BC-GP or BC-GN Panels on the BIOFIRE® (BCID2) or Luminex VERIGENE® (BC-GP, BC-GN) systems.
What if we could tackle AMR before it became a life-or-death situation?

Surveillance techniques offer an opportunity to forecast the next big resistant bug in a non-invasive and easy-to-implement way. One such method is wastewater-based epidemiology (WBE), which involves the surveillance of community wastewater samples for pathogens of interest. WBE has been around for hundreds of years — thanks John Snow (no, not that one)! The recent widespread re-implementation of WBE didn’t occur until 2020, when scientists realized WBE was a powerful tool for tracking SARS-CoV-2. The basis for SARS-CoV-2-focused WBE is simple – people infected with SARS-CoV-2 shed viral RNA in their feces, which are flushed down the toilet and begin their journey to the wastewater treatment plant, where the water will be filtered and sterilized. But if the wastewater is sampled prior to treatment, we are able to isolate that viral RNA to determine the burden of SARS-CoV-2 within a community. This became such a useful means of tracking SARS-CoV-2 burden that the Centers for Disease Control and Prevention (CDC) actually launched the National Wastewater Surveillance System (NWSS) to facilitate the tracking of SARS-CoV-2 in wastewater nationwide.

Sound interesting? Check out our webinar “What lies beneath: Wastewater testing for pathogens”
But wastewater contains more than just SARS-CoV-2 RNA. Microorganisms like bacteria, yeasts and other viruses can be tracked using WBE. Many hospitals implement a form of internal WBE during outbreaks of AMR bugs to estimate the scope of the transmission. However, by the time hospitals start to track the outbreak, it’s too late – it’s already an outbreak. In an ideal world, we would know the AMR outlook in advance, giving us the chance to intercept the culprit(s) before it becomes a hospital-wide issue.
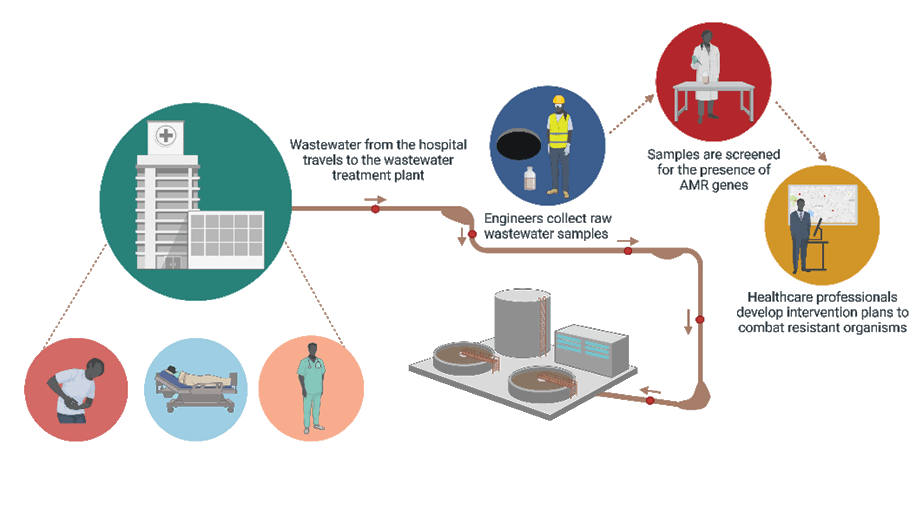
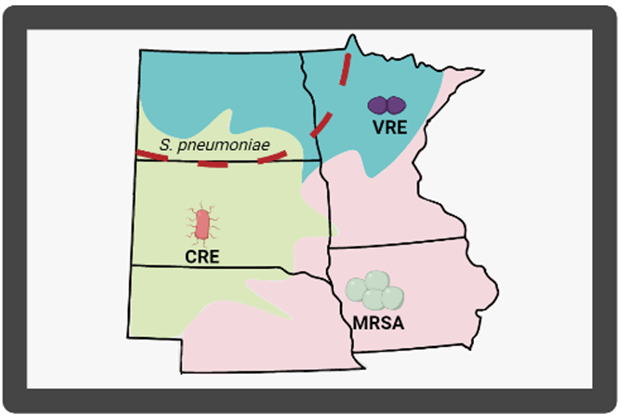
Enter community based WBE. Microorganisms can show up in community wastewater up to two weeks before patients arrive in the clinic or hospital complaining of issues. By monitoring wastewater in the community, healthcare providers could estimate the next AMR issue. Think of it like a weather report, but for microbes. Not only would this prepare providers for the influx of patients infected with resistant microbes, but it could also provide insight into the kinds of bugs that are in the community. This is an important consideration because providers could be given a short list of organisms present in the community which could help narrow down treatment options, curbing the misuse/overuse of antibiotics.
How do we know what organisms are present?
For this forecasting endeavor to be successful, a few things must happen:
- Wastewater sampling must be comprehensive. This means multiple sites statewide and nationwide must participate.
- Sample testing needs to require little time and effort.
- Nanotrap® Microbiome Particles capture and concentrate microbes in wastewater samples in less time than typical concentration methods, such as filtration or PEG precipitation.
- Analysis of samples for AMR genes must be accurate and efficient.
- Our comprehensive ARM-D® Kits test for the presence of genes encoding carbapenemases, extended-spectrum β-lactamases, plasmid-mediated ampC and other emerging threats. These kits have a high degree of sensitivity and specificity, providing you accurate, reliable results. Every kit includes positive controls for each gene target to monitor analytical process as well as an endogenous internal control (16SrRNA gene) to reduce false negatives due to PCR inhibition, DNA degradation or poor extraction. ARM-D kits are compatible with most 4-channel PCR system, making it easy to integrate them into existing procedures and protocols.
ARM-D Kits and Nanotrap Microbiome Particles are For Research Use Only. Not for use in diagnostic procedures.

AMR in Agriculture

