Abstract Control Number: 4576 | AACR 2025
Streck Research and Development, La Vista, NE
Sequential purification of cfDNA and cfRNA from blood stabilized in Nucleic Acid BCT™ using an automated workflow
Lisa Bartron MB(ASCP)CM, Jordan LaRue, and Nicholas M. George, Ph.D.
INTRODUCTION
Liquid biopsies, which interrogate body fluids in search of analytes released by healthy or diseased cells, have become increasingly common in oncology. Foremost among the technologies used are those that employ nucleic acids, such as cell-free DNA (cfDNA) and cell-free RNA (cfRNA). While cfDNA remains the primary choice for mutation detection, cfRNA is also informative, as it is released by both live and dying cells and represents the true disease state-mediated transcriptional output. Further, gene fusion events, which may be expressed at high levels in tumors, are easier to detect using cfRNA. As such, combined analysis of both cfDNA and cfRNA could provide a more comprehensive analysis. However, if blood samples are not stable, cell death or activation could lead to spurious increases in both analytes. To address this issue, Streck developed Nucleic Acid BCT, a stabilizing blood collection tube formulated to minimize changes to cfDNA and cfRNA concentrations during storage and shipping.
Nucleic Acid BCT is for Research Use Only. Not for use in diagnostic procedures. Nucleic Acid BCT should only be used for research or in the development of new assays.
MATERIALS AND METHODS
To determine whether a combined cfDNA and cfRNA isolation protocol is compatible with Nucleic Acid BCT, blood from self-declared healthy donors was drawn into EDTA and Nucleic Acid BCT and stored at ambient temperature for up to 5 days. Blood was then processed to plasma using the double-spin centrifugation protocol described in the Nucleic Acid BCT Instructions For Use (IFU). Plasma was aliquoted (4 mL per blood collection tube type and storage time) and frozen -80 °C for later bulk processing. A sequential cfDNA and cfRNA isolation protocol was completed using the Promega Maxwell® System with associated extraction kits (Maxwell® RSC ccfDNA LV Plasma Kit and Maxwell® RSC miRNA Plasma and Serum Kit, respectively) according to manufacturer’s application note (PA844, Promega Corporation). Resultant cfDNA and cfRNA were quantified using the QuantiFluor® ONE dsDNA System or QuantiFluor® RNA System, respectively, per manufacturer’s recommendations (Promega Corporation). To contrive lung cancer-positive samples, thawed plasma samples were spiked with 169-mer oligonucleotides (Integrated DNA Technologies, Inc.) harboring a EGFRL858R mutation or exon 19 deletion (E746-A750) to a target mutant allele fraction (MAF) of ~5% based on 5 ng cfDNA per mL plasma for healthy donors and a 104-mer RNA oligonucleotide (Integrated DNA Technologies, Inc.) harboring a EML4-ALK fusion junction to a target of 5% of the copy number of endogenous ALK observed in blood plasma. Spike-in nucleic concentrations were determined with ddPCR using synthesized primers and probes specific to EGFR, EGFRL858R, EGFRe19Δ, EML4-ALK (Integrated DNA Technologies, Inc.) and commercially available primer and probe for GAPDH (Applied Biosystems™). Reverse transcription of cfRNA was carried out using the iScript™ cDNA Synthesis Kit (Bio-Rad). All ddPCR reactions were run on the QX600 AutoDG™ Droplet Digital™ PCR System (Bio-Rad). Results were analyzed using GraphPad Prism 10 software (GraphPad Software).
Maxwell Systems, Maxwell RSC ccfDNA LV Plasma Kit, Maxwell RSC miRNA Plasma and Serum Kit, QuantiFluor ONE dsDNA System, and QuantiFluor RNA System are for Research Use Only. Not for use in diagnostic procedures.
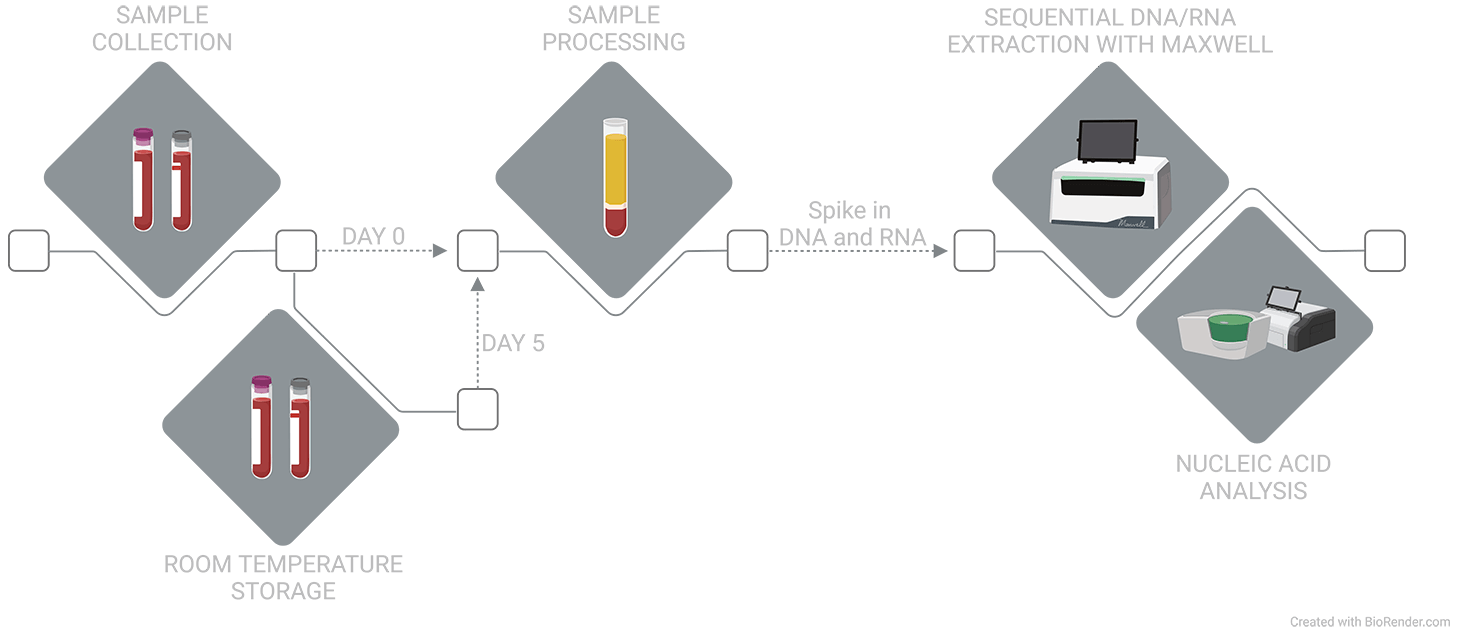
Figure 1. Blood from self-declared healthy donors was collected into EDTA or Nucleic Acid BCT. At draw or after 5 days of ambient temperature storage, plasma was isolated and frozen at -80 ˚C. Thawed plasma was spiked with DNA and RNA oligos and plasma nucleic acids were then sequentially extracted using the Promega Maxwell System and analyzed.
RESULTS
Our data support the combined use of the Promega Maxwell system with applicable extraction kits with plasma isolated from Nucleic Acid BCT. As expected, samples collected into Nucleic Acid BCT had minimal increases in cfDNA and cfRNA concentrations during storage compared to blood samples stored in EDTA. Baseline MAF of the spiked mutant cfDNA alleles (EGFRL858R and EGFRe19Δ) was completely lost in samples stored in EDTA, while equivalent blood samples stored in Nucleic Acid BCT maintained MAFs reflective of draw-time levels. A similar trend was observed when plasma RNA expression was investigated where samples collected in Nucleic Acid BCT had similar concentrations of cfRNA at draw and after storage, but those collected into EDTA did not. Specifically, EML4-ALK copy number relative to the GAPDH housekeeping gene fell precipitously in EDTA but maintained draw-time frequencies in blood samples stored in Nucleic Acid BCT.
Nucleic Acid BCT maintains draw-time plasma cell-free DNA levels and spike-in mutant allele frequencies during ambient temperature storage

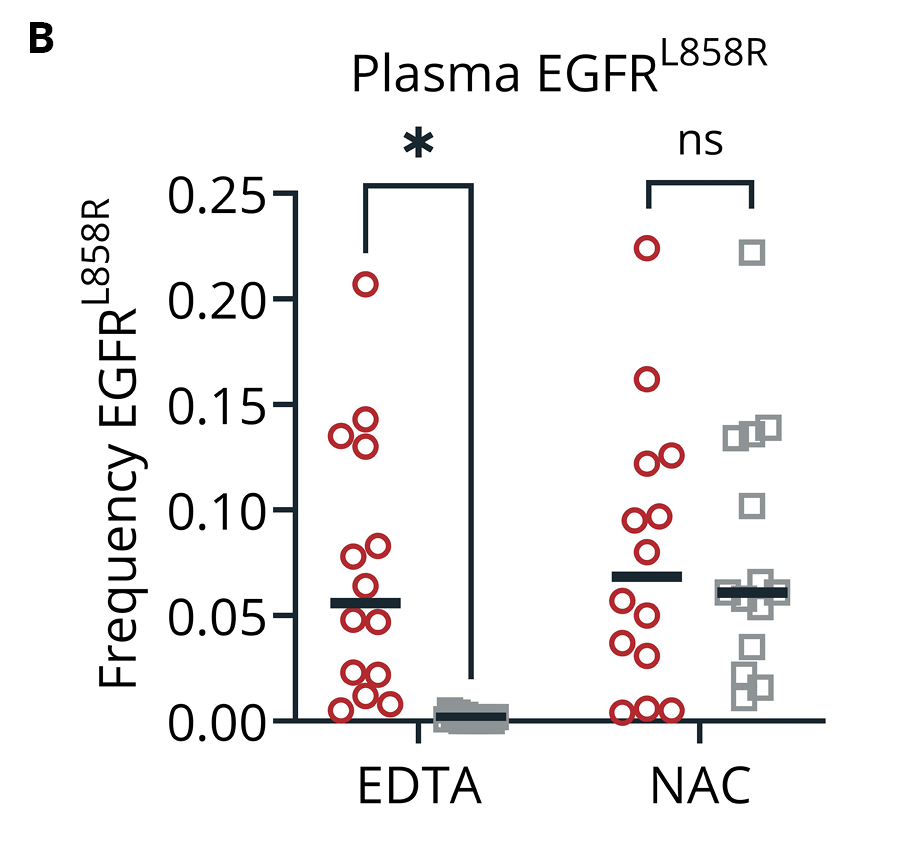

Figure 2. Concentration of total plasma DNA (A) and mutant allele frequency of EGFRL858R (B) and EGFRe19Δ (C) in samples collected into EDTA and Nucleic Acid BCT (NAC) at draw or after 5 days of ambient temperature storage (day 5). Means ± SEM are shown (n=14). *, p < 0.05 by student’s t-test; ns, not significant.
Nucleic Acid BCT maintains draw-time plasma cell-free RNA levels and spike-in fusion allele gene frequency during ambient temperature storage
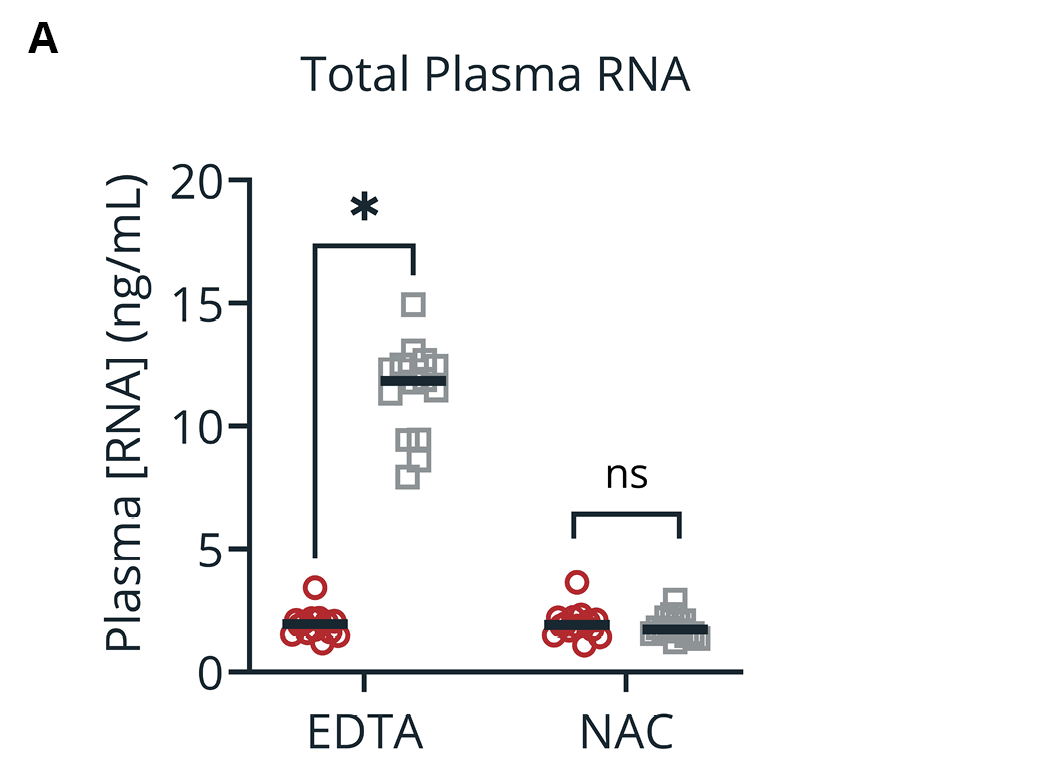

Figure 3. Concentration of total plasma RNA (A) and EML4-ALK frequency (relative to GAPDH) (B) in samples collected into EDTA and Nucleic Acid BCT (NAC) at draw or after 5 days of ambient temperature storage (day 5). Means ± SEM are shown (n=14). *, p < 0.05 by student’s t-test; ns, not significant.
CONCLUSION
Taken together, these data indicate that the combined use of Nucleic Acid BCT and the sequential nucleic acid extraction protocol described herein can provide the liquid biopsy community with a robust workflow for both blood sample stabilization and multianalyte extraction and downstream analysis.
Contact stabilization@streck.com for more information about Streck products!
Maxwell and QuantiFluor are registered trademarks of Promega Corporation.

